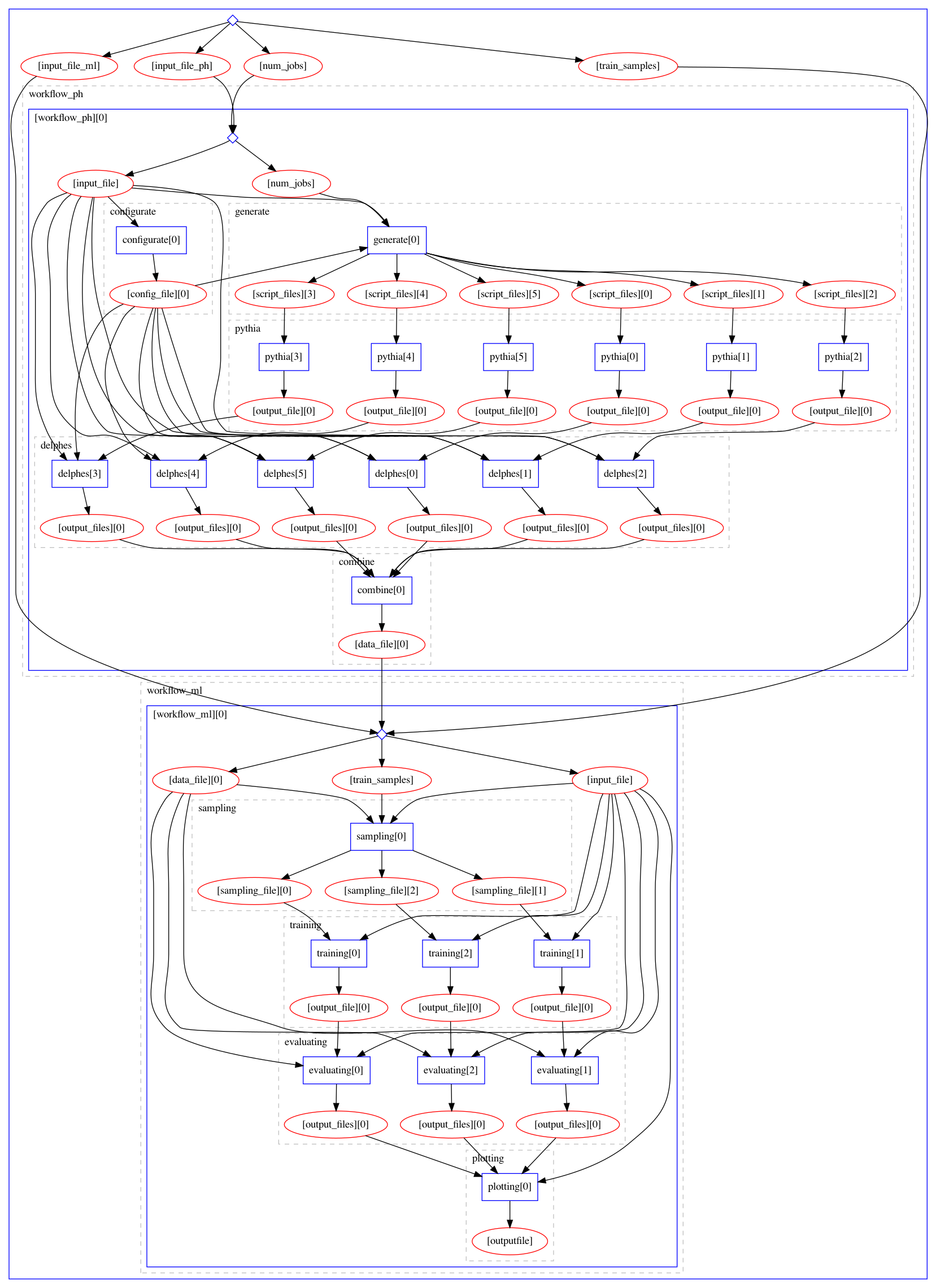
MadMiner workflow#
Public repositories#
The workflow is currently split into 3 different GitHub repositories:
Madminer-workflow-ph: containing the event generation and detector simulation part.
Madminer-workflow-ml: containing the ML training and inference part.
Madminer-workflow: wrapping both parts to construct the complete workflow.
The distribution of the workflow into 2 separate sub-workflows (PH and ML) with a wrapper around them, allows us to make both of those sub-workflow locally executable on their own. This way, researchers have the option to locally run a particular part of the complete workflow multiple times, with different parameters, without having to deal with the other part.
Setup instructions#
In order to set up the complete workflow locally, you can just clone the madminer-workflow repository, as long as the two wrapped sub-workflows by running:
git clone --recurse-submodules https://github.com/madminer-tool/madminer-workflow
This command will also clone the Physics (PH) and Machine Learning (ML) sub-workflows inside the modules folder.
Optionally, you could install them separately (if you want to interact with them in a separate manner), by running:
git clone https://github.com/madminer-tool/madminer-workflow-ml
git clone https://github.com/madminer-tool/madminer-workflow-ph
The default example#
These repositories define a default workflow as an example, but you could modify the configuration files to change either the Physics process that you are interested, or the ML models / inference strategies you would like to use. In the section titled Parametrization, we describe how to modify the workflow’s configuration files for your own research purposes. You can skip that for now and go straight to the following section.
The image below shows the default workflow structure