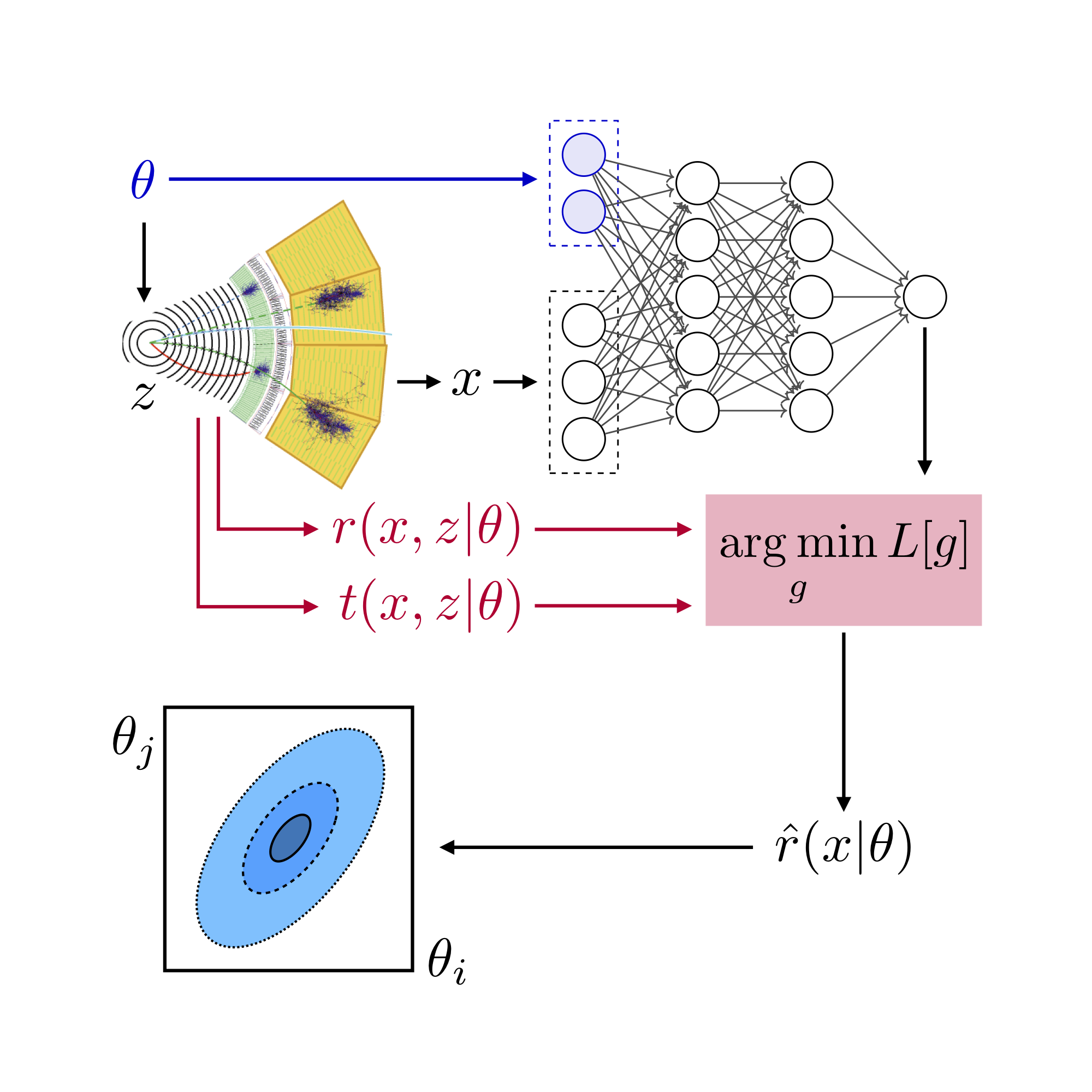
Appendix 1: Adding systematic uncertainties#
Johann Brehmer, Felix Kling, Irina Espejo, and Kyle Cranmer 2018-2019
In this tutorial we’ll explain how to add systematic uncertainties to the MadMiner workflow.
Preparations#
Before you execute this notebook, make sure you have MadGraph, Pythia and Delphes installed.
import os
import logging
import numpy as np
import matplotlib
from matplotlib import pyplot as plt
%matplotlib inline
from madminer.core import MadMiner
from madminer.lhe import LHEReader
from madminer.sampling import SampleAugmenter
from madminer import sampling
from madminer.ml import ScoreEstimator
from madminer.fisherinformation import FisherInformation, profile_information, project_information
from madminer.plotting import plot_systematics, plot_fisher_information_contours_2d
Please enter here the environment variable pointing to your MG5 installation folder.
mg_dir = os.getenv('MG_FOLDER_PATH')
MadMiner uses the Python logging module to provide additional information and debugging output. You can choose how much of this output you want to see by switching the level in the following lines to logging.DEBUG or logging.WARNING.
# MadMiner output
logging.basicConfig(
format='%(asctime)-5.5s %(name)-20.20s %(levelname)-7.7s %(message)s',
datefmt='%H:%M',
level=logging.DEBUG
)
# Output of all other modules (e.g. matplotlib)
for key in logging.Logger.manager.loggerDict:
if "madminer" not in key:
logging.getLogger(key).setLevel(logging.WARNING)
1. Parameters and benchmarks#
We’ll just load the MadMiner setup from the first part of this tutorial:
miner = MadMiner()
miner.load('data/setup.h5')
16:19 madminer.utils.inter DEBUG HDF5 file does not contain is_reference field.
16:19 madminer.core INFO Found 2 parameters:
16:19 madminer.core INFO CWL2 (LHA: dim6 2, maximal power in squared ME: (2,), range: (-20.0, 20.0))
16:19 madminer.core INFO CPWL2 (LHA: dim6 5, maximal power in squared ME: (2,), range: (-20.0, 20.0))
16:19 madminer.core INFO Found 6 benchmarks:
16:19 madminer.core INFO sm: CWL2 = 0.00e+00, CPWL2 = 0.00e+00
16:19 madminer.core INFO w: CWL2 = 15.20, CPWL2 = 0.10
16:19 madminer.core INFO neg_w: CWL2 = -1.54e+01, CPWL2 = 0.20
16:19 madminer.core INFO ww: CWL2 = 0.30, CPWL2 = 15.10
16:19 madminer.core INFO neg_ww: CWL2 = 0.40, CPWL2 = -1.53e+01
16:19 madminer.core INFO morphing_basis_vector_5: CWL2 = -1.58e+01, CPWL2 = -1.94e+01
16:19 madminer.core INFO Found morphing setup with 6 components
16:19 madminer.core INFO Did not find systematics setup.
2. Set up systematics, save settings#
This is where things become interesting: We want to model systematic uncertainties. The main function is add_systematics(), the keyword effect determines how the effect of the nuisance parameters on the event weights is calculated. For effect="norm", the nuisance parameter rescales thee overall cross section of one or multiple samples. For effect="pdf", its effect is calculated with PDF variations. Finally, with effect="scale" scale variations are used.
Here we consider three nuisance parameters: one for the signal normalization, one for the background normalization, and one for scale uncertainties (which we here assume to be correlated between signal and background).
miner.add_systematics(effect="norm", systematic_name="signal_norm", norm_variation=1.1)
miner.add_systematics(effect="norm", systematic_name="bkg_norm", norm_variation=1.2)
miner.add_systematics(effect="scale", systematic_name="scales", scale="mu")
Again, we save our setup:
miner.save('data/setup_systematics.h5')
16:19 madminer.core INFO Saving setup (including morphing) to data/setup_systematics.h5
3. Run MadGraph#
Now it’s time to run MadGraph. MadMiner will instruct MadGraph to use its built-in systematics tool to calculate how the event weights change under the scale variation.
miner.run(
sample_benchmark='sm',
mg_directory=mg_dir,
mg_process_directory='./mg_processes/signal_systematics',
proc_card_file='cards/proc_card_signal.dat',
param_card_template_file='cards/param_card_template.dat',
run_card_file='cards/run_card_signal_small.dat',
log_directory='logs/signal',
python2_override=True,
systematics=["signal_norm", "scales"],
)
16:20 madminer.utils.inter INFO Generating MadGraph process folder from cards/proc_card_signal.dat at ./mg_processes/signal_systematics
16:20 madminer.core INFO Run 0
16:20 madminer.core INFO Sampling from benchmark: sm
16:20 madminer.core INFO Original run card: cards/run_card_signal_small.dat
16:20 madminer.core INFO Original Pythia8 card: None
16:20 madminer.core INFO Original config card: None
16:20 madminer.core INFO Copied run card: /madminer/cards/run_card_0.dat
16:20 madminer.core INFO Copied Pythia8 card: None
16:20 madminer.core INFO Copied config card: None
16:20 madminer.core INFO Param card: /madminer/cards/param_card_0.dat
16:20 madminer.core INFO Reweight card: /madminer/cards/reweight_card_0.dat
16:20 madminer.core INFO Log file: run_0.log
16:20 madminer.core INFO Creating param and reweight cards in ./mg_processes/signal_systematics//madminer/cards/param_card_0.dat, ./mg_processes/signal_systematics//madminer/cards/reweight_card_0.dat
16:20 madminer.utils.inter INFO Starting MadGraph and Pythia in ./mg_processes/signal_systematics
16:23 madminer.core INFO Finished running MadGraph! Please check that events were succesfully generated in the following folders:
./mg_processes/signal_systematics/Events/run_01
miner.run(
sample_benchmark='sm',
mg_directory=mg_dir,
mg_process_directory='./mg_processes/bkg_systematics',
proc_card_file='cards/proc_card_background.dat',
param_card_template_file='cards/param_card_template.dat',
run_card_file='cards/run_card_background.dat',
log_directory='logs/background',
python2_override=True,
systematics=["bkg_norm", "scales"],
)
16:23 madminer.utils.inter INFO Generating MadGraph process folder from cards/proc_card_background.dat at ./mg_processes/bkg_systematics
16:23 madminer.core INFO Run 0
16:23 madminer.core INFO Sampling from benchmark: sm
16:23 madminer.core INFO Original run card: cards/run_card_background.dat
16:23 madminer.core INFO Original Pythia8 card: None
16:23 madminer.core INFO Original config card: None
16:23 madminer.core INFO Copied run card: /madminer/cards/run_card_0.dat
16:23 madminer.core INFO Copied Pythia8 card: None
16:23 madminer.core INFO Copied config card: None
16:23 madminer.core INFO Param card: /madminer/cards/param_card_0.dat
16:23 madminer.core INFO Reweight card: /madminer/cards/reweight_card_0.dat
16:23 madminer.core INFO Log file: run_0.log
16:23 madminer.core INFO Creating param and reweight cards in ./mg_processes/bkg_systematics//madminer/cards/param_card_0.dat, ./mg_processes/bkg_systematics//madminer/cards/reweight_card_0.dat
16:23 madminer.utils.inter INFO Starting MadGraph and Pythia in ./mg_processes/bkg_systematics
4. Load events from LHE file#
When adding LHE or Delphes files, use the systematics keyword to list which systematic uncertainties apply to which sample:
lhe = LHEReader('data/setup_systematics.h5')
lhe.add_sample(
lhe_filename='mg_processes/signal_systematics/Events/run_01/unweighted_events.lhe.gz',
sampled_from_benchmark='sm',
is_background=False,
k_factor=1.1,
systematics=["signal_norm", "scales"]
)
lhe.add_sample(
lhe_filename='mg_processes/bkg_systematics/Events/run_01/unweighted_events.lhe.gz',
sampled_from_benchmark='sm',
is_background=True,
k_factor=1.1,
systematics=["bkg_norm", "scales"]
)
The next steps are unaffected by systematics.
lhe.set_smearing(
pdgids=[1,2,3,4,5,6,9,21,-1,-2,-3,-4,-5,-6], # Partons giving rise to jets
energy_resolution_abs=0.,
energy_resolution_rel=0.1,
pt_resolution_abs=None,
pt_resolution_rel=None,
eta_resolution_abs=0.1,
eta_resolution_rel=0.,
phi_resolution_abs=0.1,
phi_resolution_rel=0.,
)
lhe.add_observable(
'pt_j1',
'j[0].pt',
required=False,
default=0.,
)
lhe.add_observable(
'delta_phi_jj',
'j[0].deltaphi(j[1]) * (-1. + 2.*float(j[0].eta > j[1].eta))',
required=True,
)
lhe.add_observable(
'met',
'met.pt',
required=True,
)
lhe.add_cut('(a[0] + a[1]).m > 122.')
lhe.add_cut('(a[0] + a[1]).m < 128.')
lhe.add_cut('pt_j1 > 30.')
lhe.analyse_samples()
lhe.save('data/lhe_data_systematics.h5')
A look at distributions#
The function plot_systematics() makes it easy to check the effect of the various nuisance parameters on a distribution:
plot_systematics?
_ = plot_systematics(
filename='data/lhe_data_systematics.h5',
theta=np.array([0.,0.]),
observable="pt_j1",
obs_label="$p_{T,j}$",
obs_range=(30.,400.),
ratio_range=(0.7,1.4)
)
_ = plot_systematics(
filename='data/lhe_data_systematics.h5',
theta=np.array([0.,0.]),
observable="delta_phi_jj",
obs_label=r"$\Delta\phi$",
obs_range=(-3.1,3.1),
ratio_range=(0.7,1.4)
)
5. Sampling#
Let’s generate training data for the SALLY method.
sampler = SampleAugmenter('data/lhe_data_systematics.h5', include_nuisance_parameters=True)
x, theta, t_xz, _ = sampler.sample_train_local(
theta=sampling.benchmark('sm'),
n_samples=100000,
folder='./data/samples',
filename='train_score_systematics'
)
6. Training#
The SALLY estimator will learn the score in terms of both the physics parameters and the nuisance parameters. In our case, its output will therefore be a vector with five components.
estimator = ScoreEstimator(n_hidden=(50,))
estimator.train(
method='sally',
x='data/samples/x_train_score_systematics.npy',
t_xz='data/samples/t_xz_train_score_systematics.npy',
)
estimator.save('models/sally_systematics')
7. Fisher information#
The Fisher information is now also a 5x5 matrix. Constraint terms on the nuisance parameters, representing our prior knowledge on their values, can be calculated with FisherInformation.nuisance_constraint_information().
fisher = FisherInformation('data/lhe_data_systematics.h5')
fisher.full_information?
info_sally, _ = fisher.full_information(
theta=np.zeros(5),
model_file='models/sally_systematics',
luminosity=1000000.,
include_xsec_info=False,
)
info_nuisance = fisher.nuisance_constraint_information()
info = info_sally + info_nuisance
for row in info:
print(" ".join(["{:6.2f}".format(entry) for entry in row]))
Fisher contours and profiled information#
When looking at a subset of these five parameter, we can either ignore the other parameters (“project”), or conservatively pick the most pessimistic value of them (“profile” them). MadMiner provides the functions profile_information() and project_information() for this purpose. Let’s do that for the parameter space of the two physics parameters:
info_proj = project_information(info, [0,1])
info_prof = profile_information(info, [0,1])
_ = plot_fisher_information_contours_2d(
[info_proj, info_prof],
inline_labels=["Projected", "Profiled"],
xrange=(-0.1,0.1),
yrange=(-0.1,0.1)
)
For illustration, we can look at just one physics parameter and one nuisance parameter:
info_demo = project_information(info, [0, 4])
info_demo_profiled = np.zeros((2,2))
info_demo_profiled[0,0] = profile_information(info_demo, [0,])
_ = plot_fisher_information_contours_2d(
[info_demo, info_demo_profiled],
inline_labels=[None, "Profiled"],
xrange=(-0.2,0.2),
yrange=(-0.6,0.6),
xlabel="Parameter of interest",
ylabel="Nuisance parameter",
)
plot_fisher_information_contours_2d?